
Unsere Arbeitsgruppe nutzt "Genome Editing" zur Entwicklung von Nutzpflanzen wie Reis, Kartoffel, Erdnuss oder Senf, mit verbesserten Eigenschaften. Dazu entwickeln wir neue TALEN- und CRISPR-Werkzeuge und erforschen die Strategie pflanzlicher Schädlinge.
Bakterielle Pflanzenpathogene und ihre Waffen
Zwei Organismen kämpfen ums Überleben. Der Prokaryot zielt darauf ab, Nährstoffe zu erhalten, während der Eukaryot darauf abzielt, die Integrität seiner Zellen zu schützen. Pflanzen sind durchaus in der Lage, sich zu verteidigen, deshalb können nur spezialisierte Pathogene sie kolonisieren.
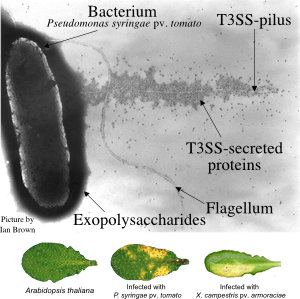
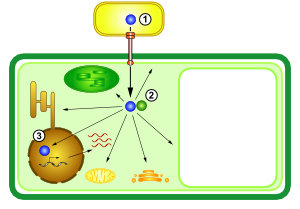
Diese Pathogene haben ausgeklügelte molekulare Waffen entwickelt. Pseudomonas syringae und Xanthomonas-Bakterien verwenden ein Typ-III-Sekretionssystem (T3SS), um ca. 30 verschiedene Effektorproteine in Pflanzenzellen zu injezieren (Abb. 1). Diese Effektoren manipulieren die Wirtszelle von innen heraus (Abb. 2). In unserer Arbeitsgruppe untersuchen wir ihre molekularen Aktivitäten.
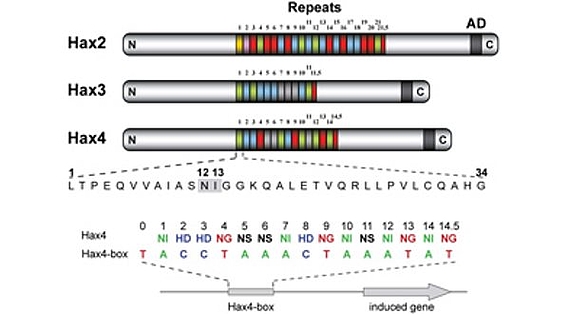
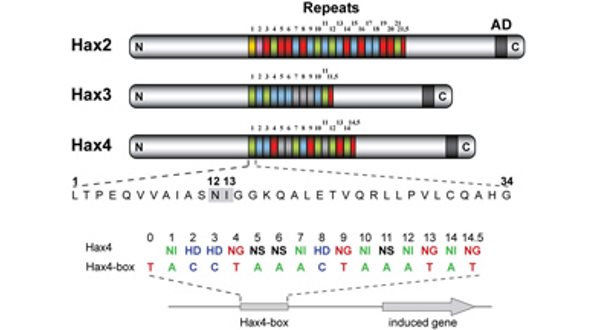
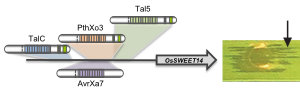
TALEs (Transcription Activator-Like Effectors) reispathogener Xanthomonas oryzae induzieren pflanzliche SWEET-Zuckerexporter. X. oryzae-Stämme können bis zu 26 verschiedene TALEs besitzen. In diesem Zusammenhang erforschen wir, welche Pflanzengene die Ziele dieser TALEs sind (Abb. 3)






Leitung


30419 Hannover




